You are using a web browser that is not fully supported by this website. Some features may not work as intended. For the best experience, please use one of the recommended browsers.



Opening image: RNA pseudoknot from simian retrovirus type-1 (SRV-1).
RNA pseudoknots are found in virtually all classes of RNA and play key roles in a variety of biological processes. The ability to predict pseudoknots computationally from sequence data is quite limited because no precise rules exist, especially for complex pseudoknots which contain pseudoknots within pseudoknots. Experimental evidence for pseudoknots in RNA molecules comes from sequence comparisons, along with nuclease digestion, chemical modification, and NMR studies.
In this tutorial we'll explore Juli Feigon's recent structural and thermodynamic studies of the wild-type and mutant human telomerase pseudoknot. This exciting work has provided the first 3D structural insight into the molecular basis of a human disease caused by a defect in an RNA pseudoknot.
Pseudoknots are required for the replication of retroviruses. Therefore, the pseudoknot structures in the viral RNA genome are attractive targets for the development of antiviral drugs.
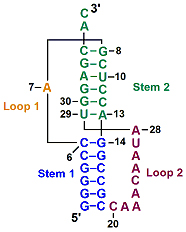
A simple pseudoknot is composed of two short helical segments connected by single-stranded regions or loops (). The two stems are able to stack on top of each other to form a quasi-continuous helix with one continuous strand and one discontinuous strand. Complex pseudoknots may contain pseudoknots within the loops.
The stack on top of each other to form a quasi-continuous helix. One strand (8-19) is continuous, the other discontinuous (see ).
spacefill. Describe how the single-stranded loops interact with the adjacent stems.
Telomeres are highly organized protein-nucleic complexes which are located at the ends of eukaryotic chromosomes. Telomeres are unstable due to the loss of telomeric DNA at each cell division, until a critically short length occurs, causing cell cycle arrest or apoptosis. Learn more.
Telomerase RNA contains a pseudoknot. Human telomerase contains a 451 nucleotide RNA (hTR) and several proteins, including a specialized reverse transcriptase. The 5′ half of hTR comprises the pseudoknot domain, which includes the RNA template for telomere synthesis and a highly conserved pseudoknot that is required for telomerase activity.
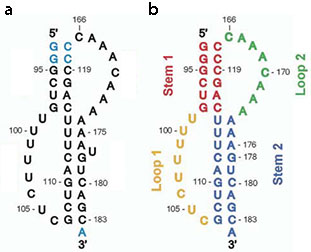
A unimolecular pseudoknot construct composed of nucleotides 93-121 and 166-184 with a U177 deletion was used for the thermodynamic and NMR studies ().
The NMR structure of this essential pseudoknot reveals a well-defined 3D structure with extensive loop-loop and loop-stem interactions.
spacefill.
The adenine-rich Loop 2 (green) sits in the minor groove of Stem 1 (red), while the uracil-rich Loop 1 (yellow) fits into the major groove of Stem 2 (blue). Describe the loop-loop interaction.
Normally, the two stems of a pseudoknot are stacked coaxially. However, the 15-bp helix in the ΔU177 pseudoknot is slightly bent compared to the other 15-bp helix.
The A173·U99 at the junction of the red and blue helices precludes a coaxial alignment of the two helices.
A triple helix stabilizes the pseudoknot and is essential for activity.
Five important base triples flank the helical junction and form a continuous triple helix. A triple helix within the pseudoknot is a conserved and essential element of telomerase RNA. While triplex-disrupting mutations abolish telomerase function, triple mutations that form isosteric triples restore the pseudoknot structure and restore telomerase function in vivo.
Five nucleotides, shown in CPK colors, from the two loops are nested in the grooves of the helical stems. Each of these five bases forms a base triple by hydrogen bonding to the edges of Watson-Crick base pairs in the helix stems.
The U115·A174(U100) base triple (the third base is enclosed in parentheses). As expected, all three bases lie in a plane. spacefill.
all five base triples (CPK colors). The presence of the conserved A·U base pair (purple) in the triple helix is puzzling (see below). spacefill.
The tertiary interactions between five base triples create a triple helix which makes a significant contribution to the stability of the pseudoknot. Although the existence of triple helices has been known for decades, this is the first triple helix in a natural RNA for which the 3D strucure has been determined.
After looking at the NMR structure of the ΔU177 pseudoknot, you're probably wondering whether the WT pseudoknot has a similar structure. Wouldn't the U177 bulge affect the way the WT pseudoknot folds?
The answer is yes. A176 and G178 (blue) are in Stem 2 and an unpaired base at position 177 would distort the helix. Thus, one would be expect small differences in the 3D structures of the WT and ΔU177 pseudoknots. However, since the U177 bulge is conserved, it must make an important contribution to the structure and function of the telomerase pseudoknot.
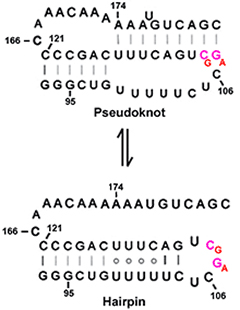
Feigon's group was able to solve the NMR structure of the ΔU177 pseudoknot because deletion of U177 bulge stabilizes the pseudoknot relative to an alternate conformation, namely a hairpin (). Conversely, the U177 bulge destabilizes the pseudoknot relative to the hairpin conformation and the two conformations are in equilibrium. For this reason, the NMR structure of the pseudoknot cannot be determined unless one deletes U177.
NMR structure of a 30-nucleotide construct containing residues 93-121 and 166. The hairpin has a well-defined structure consisting of a 12-bp helix capped by a pentaloop.
The stem contains three consecutive U·U base pairs stacked on six Watson-Crick pairs, followed by a U·C pair and two additional Watson-Crick base pairs (). The nucleotides that form the "uracil helix" in the hairpin are 100% conserved in vertebrates. However, conservation of the U99→U102 sequence is not required for the pseudoknot.
the uracil helix. This is the only example of four consecutive uridines in the various RNA databases.
Mutations in telomerase RNA pseudoknot cause dyskeratosis congenita (DKC), a rare, progressive bone marrow disease. Patients with DKC have reduced telomerase activity and abnormally short tracts of telomeric DNA. A monogenetic disease, DKC is caused by mutations in either dyskerin, a key protein in the telomerase complex, or the telomerase RNA pseudoknot. Thermodynamic and structural studies of pseudoknot constructs have shown that mutations linked to DKC cause changes in the equilibrium of telomerase RNA pseudoknot and hairpin conformers.
The bases G107 and C108 of the GC→AG double mutant are located in the pentaloop of the hairpin, where they would be expected to have a minimal impact on its stability. Explain why these mutations would be expected to destabilize the pseudoknot structure and thus shift the equilibrium toward the hairpin.