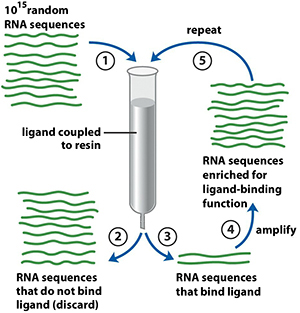
Selection of Aptamers Using The SELEX Method
Basic steps in the SELEX selection process are presented in Fig. 1. Over 1015 different DNA oligonucleotides can be created by programming a DNA synthesizer to add equal amounts of G, A, T, and C phosphoramidites at each step of the synthesis. For the selection of DNA aptamers, this library can be used directly in the first round of a DNA SELEX process. For the selection of RNA aptamers, the random DNA oligonucleotide library has to be transformed into a RNA library before starting the first round of an RNA SELEX process. In essence, these repetitive cycles of in vitro selection and enzymatic amplification mimic Darwinian evolution, only that "survival-of-the-fittest" is carried out at the level of individual molecules as opposed to individual organisms. SELEX drives the selection towards a few structures optimized for binding a specific ligand.