π-Stacking of Aromatic Side Chains in Proteins
Over thirty thousand pairs of π-π interactions between aromatic side chains of proteins have been found in the Protein Data Bank. The observed orientations can be explained on the basis of a simple electrostatic model for the energy of interaction (see Fig. 1).
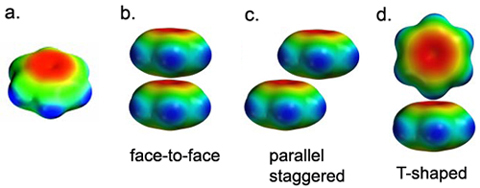
Figure 1.A shows an electrostatic potential map for benzene. Note that the surface is colored red in the π system (indicating negative potential and the fact that this region is attracted to a positive charge), and blue in the σ system (indicating positive potential and the fact that this region is repelled by a positive charge). If two phenylalanine rings are stacked on top of one another, the repulsion due to the negatively charged π-electron clouds outweighs any attractive forces (Fig. 1.B). In fact, face-to-face stacking of Phe rings is not observed in proteins. Even in the nucleic acids, where "stacking" is traditionally invoked as one of the forces that stabilizes the double helical structure, true face-to-face stacking rarely occurs. Usually the bases are staggered or twisted away from face-to-face geometry.
The most common geometry for π-stacking of aromatic side chains is the T-shaped or edge-to-face orientation (Fig. 1.D). In this orientation, the positively charged framework of one ring is juxtaposed against the π electron cloud of another ring. Favorable interactions also occur if the rings are parallel, but staggered (Fig. 1.C).